For onsite builds at all levels, you do not find out the specific section until you arrive at the competition. You might be able to make an educated guess based on the location of important structural features, but I would be familiar with the whole thing.jkatz16 wrote:Does anyone know what the restriction numbers are for the onsite build for states, the TAL effectors? Thanks
Protein Modeling C
-
Gemma W
- Member

- Posts: 196
- Joined: Thu Mar 10, 2011 2:12 pm
- Division: Grad
- State: NY
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
2015 events: WIDI, Protein Modeling, Geomapping, Chem Lab
2014 events: WIDI, Geomapping, Materials Science, Food Science
2013 events: WIDI, Mousetrap Vehicle, Heredity, Food Science, Metric Mastery
Best ever place: Nationals, 3rd in WIDI
2014 events: WIDI, Geomapping, Materials Science, Food Science
2013 events: WIDI, Mousetrap Vehicle, Heredity, Food Science, Metric Mastery
Best ever place: Nationals, 3rd in WIDI
-
annaphase
- Member

- Posts: 10
- Joined: Sun Feb 01, 2015 9:40 pm
- Division: C
- State: NY
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
The point is that we don't know, so we can't practice until we get therejkatz16 wrote:Does anyone know what the restriction numbers are for the onsite build for states, the TAL effectors? Thanks
"What are the squiggly things for?"~Protein Modeling
"150 degrees Celsius? Either this graph is wrong, or the only place this enzyme is active is in pigeon." ~Cell Bio
"150 degrees Celsius? Either this graph is wrong, or the only place this enzyme is active is in pigeon." ~Cell Bio
-
Gemma W
- Member

- Posts: 196
- Joined: Thu Mar 10, 2011 2:12 pm
- Division: Grad
- State: NY
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
So on the CBM website (as well as in other sources) it talks about the TALE proteins as having two alpha helices connected by a loop, and the two hypervariable residues that are contained in the loop and read DNA. However, in the picture and in the Jmol visualization of the protein, the residues in question seem to be at the end of one helix. What loop are they talking about?
2015 events: WIDI, Protein Modeling, Geomapping, Chem Lab
2014 events: WIDI, Geomapping, Materials Science, Food Science
2013 events: WIDI, Mousetrap Vehicle, Heredity, Food Science, Metric Mastery
Best ever place: Nationals, 3rd in WIDI
2014 events: WIDI, Geomapping, Materials Science, Food Science
2013 events: WIDI, Mousetrap Vehicle, Heredity, Food Science, Metric Mastery
Best ever place: Nationals, 3rd in WIDI
-
colorado mtn science
- Member

- Posts: 22
- Joined: Thu Jan 09, 2014 9:16 am
- Division: C
- State: CO
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
A beta bridge is a beta sheet (made from two beta strands connected with H bonds) where each beta strand is one residue long._vanemchir_ wrote:hey guys just a quick question... what exactly is a beta bridge and how do you fold a beta bridge on the toober? i have looked this up for a while and can't seem to find anything.
Thanks,
Vane
-
Celestite

- Member

- Posts: 13
- Joined: Sat Mar 15, 2014 1:46 pm
- Division: C
- State: NJ
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
They're talking about the tip of the loop connecting the alpha helices.Gemma W wrote:So on the CBM website (as well as in other sources) it talks about the TALE proteins as having two alpha helices connected by a loop, and the two hypervariable residues that are contained in the loop and read DNA. However, in the picture and in the Jmol visualization of the protein, the residues in question seem to be at the end of one helix. What loop are they talking about?
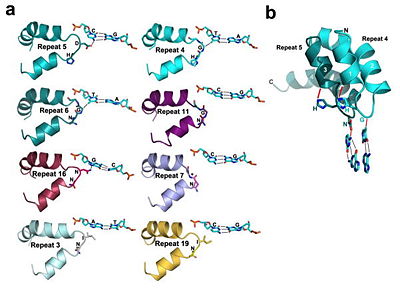
You can see it more clearly in this picture.
-
ellpida13
- Member

- Posts: 2
- Joined: Sat Feb 21, 2015 2:06 pm
- Division: C
- State: PA
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
Hey guys, I've noticed that there seems to be two sets of the FokI protein when I download the pdb file into Jmol (see the attached picture of 2fok A selected / restricted, with cartoon on and backbone at 1.5).
Can anyone explain to me why this occurs? If this is correct, do we need to reflect the second set in our pre-build model?
Thanks!
Can anyone explain to me why this occurs? If this is correct, do we need to reflect the second set in our pre-build model?
Thanks!
You do not have the required permissions to view the files attached to this post.
-
fozendog

- Member

- Posts: 193
- Joined: Tue Apr 17, 2012 5:51 pm
- Division: Grad
- State: WA
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
Restrict it to chain A with the command:
restrict *A
That is all you need for the base pre-build model.
restrict *A
That is all you need for the base pre-build model.
Stanford '19
Camas Science Olympiad Alumnus
Events: Protein Modeling, Cell Biology, Disease Detectives, Experimental Design, Dynamic Planet, Water Quality
Camas Science Olympiad Alumnus
Events: Protein Modeling, Cell Biology, Disease Detectives, Experimental Design, Dynamic Planet, Water Quality
-
bernard

- Administrator

- Posts: 2416
- Joined: Sun Jan 05, 2014 3:12 pm
- Division: Grad
- State: WA
- Pronouns: He/Him/His
- Has thanked: 179 times
- Been thanked: 759 times
Re: Protein Modeling C
If you want to do it all in one command, "restrict 421-560:A"fozendog wrote:Restrict it to chain A with the command:
restrict *A
That is all you need for the base pre-build model.
"One of the ways that I believe people express their appreciation to the rest of humanity is to make something wonderful and put it out there." – Steve Jobs
-
ellpida13
- Member

- Posts: 2
- Joined: Sat Feb 21, 2015 2:06 pm
- Division: C
- State: PA
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
I see what I was doing wrong now. Thank you fozendog and bernard for the help!
-
Gearbox
- Member

- Posts: 40
- Joined: Tue Mar 20, 2012 10:47 am
- Division: C
- State: PA
- Has thanked: 0
- Been thanked: 0
Re: Protein Modeling C
I know this is a bit of a reach, but is anyone here from Huron High School in Ann Arbor, they were recently at the Mentor Invitational and their protein was freaking amazing, especially with the copper DNA model that they built. I was wondering if anyone from that team could give me some pointers on how they built the DNA. Thanks!
2015 Events
Protein Modeling
Cell Biology
Disease Detectives
Chem Lab

Protein Modeling
Cell Biology
Disease Detectives
Chem Lab


